RESEARCH
We are interested in studying the natural systems which shape and guide the processes of the natural world. Our long-term goal is to identify and characterize the scientific mechanisms specific to our principal areas of research. You can read on to find out more about these projects below.
Protein-protein interactions play a central role in various aspects of the structural and functional organization of the cell, and their elucidation is crucial for a better understanding of processes such as metabolic control, signal transduction, and gene regulation. Genome-wide proteomics studies, primarily yeast two-hybrid assays, will provide an increasing list of interacting proteins, but only a small fraction of the potential complexes will be amenable to direct experimental analysis. Thus, it is important to develop docking methods that can elucidate the details of specific interactions at the atomic level.

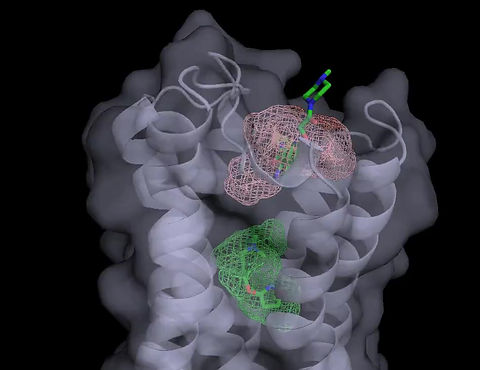
Docking small molecules to a protein is a fundamental step in structure-based drug design. The main approaches are (A) Docking of potential ligands from a compound database, and (B) mapping the protein for the binding sites of molecular probes - small molecules and functional groups - and using the favorable positions for the construction of larger ligands. We develop and apply algorithms for both approaches.
The most successful protein structure prediction method to date is homology modeling (also known as comparative modeling). The approach is based on the structural conservation of the framework regions between the members of a protein family. Since the 3D structures are more conserved in evolution than sequence, even the best sequence alignment methods frequently fail to correctly identify the regions that possess the desired level of structural similarity, and the quality of alignment is the single most important factor determining the accuracy of the 3D model.

